陶士珩
陶士珩
基本信息
陶士珩,男,教授,博士生导师。
教育经历
1978年-1982年,本科,南京气象学院农业气象专业;
1982年-1985年,硕士,西南农业大学农业气候专业;
1998年7月,博士,西北农业大学农学专业;
1999年6月-2001年5月,复旦大学遗传学研究所做群体与数量遗传学博士后研究;
2007年1月-2008年1月,赴美国亚利桑那大学进化与生态学系做访问学者研究;
工作经历
1985年,西北农业大学,从事农业气象学教学与科研;
1996年7月,西北农业大学,副教授;
2001年12月,西北农林科技大学,教授;
现任教于西北农林科技大学生命科学学院生物信息学教研组,西北农林科技大学生物信息学研究中心主任。
荣誉获奖
省部级科技进步二等奖(第三名)
国家科技进步二等奖(集体奖)
主持项目
主持国家自然科学基金1项
教育部博士点基金1项
主持国家重点实验室开放课题3项
参与“973”项目和“863”项目各1项
学科建设
建立西北农林科技大学生物信息学研究中心
课程建设
主编了《生物信息学》教材;
先后承担本科生《生物信息学》、《进化生物学》、硕士研究生《遗传分析》、《高级生物信息学》、《分子进化》、博士研究生《生物信息学研究进展》等课程的教学。
科学研究
近年来致力于生物信息学、分子进化、群体遗传学等领域的教学与科研工作。目前主要从事RNA结构调控蛋白质翻译、基因水平转移规律、互作蛋白质协同进化等领域的研究。
1. 主要研究方向
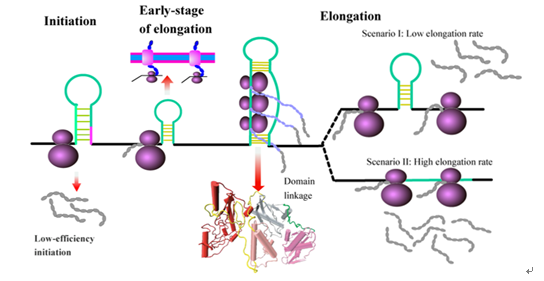
(1) RNA结构调控蛋白质翻译
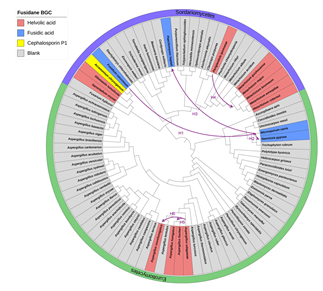
(2) 基因水平转移规律
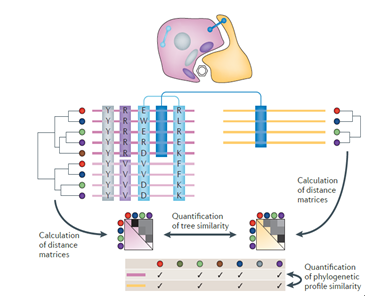
(3) 蛋白质协同进化
2. 主要学术成果
(1) 数据库与在线软件
RSVdb:转录组体内mRNA结构综合数据库(https://taolab.nwsuaf.edu.cn/rsvdb/)
CRISPR-CBEI:一个基于网络的胞嘧啶碱基编辑器介导的基因失活设计工具(https://taolab.nwsuaf.edu.cn/CrisprCBEI/)
(2) 学术论文
Yu H , Wu Z , Chen X , et al. CRISPR-CBEI: a Designing and Analyzing Tool Kit for Cytosine Base Editor-Mediated Gene Inactivation Downloaded from[J]. mSystems, 2020, 5(5).
Xiangchen L , Jian C , Xiaonan L , et al. Origin and Evolution of Fusidane-Type Antibiotics Biosynthetic Pathway through Multiple Horizontal Gene Transfers[J]. Genome Biology and Evolution(10):10.
Li Q , Chang Y , Zhang K , et al. Implication of the gut microbiome composition of type 2 diabetic patients from northern China[J]. Scientific Reports, 2020, 10(1).
Yao X , Fan Q , Yao B , et al. Codon Usage Bias Analysis of Bluetongue Virus Causing Livestock Infection[J]. Frontiers in Microbiology, 2020, 11:655.
Hua Y , Zhao Y G , Tao S H , et al. Morphogenesis of embryonic appendages of Arge pagana Panzer (Hymenoptera: Argidae)[J]. Zoologischer Anzeiger, 2020, 287.
Haopeng Y , Yi Z , Qing S , et al. RSVdb: a comprehensive database of transcriptome RNA structure[J]. Briefings in Bioinformatics.
HUA Y , ZHANG B-B , MIAO Y , et al. Vasa deferentia and associated structures of the male Panorpodes kuandianensis (Mecoptera: Panorpodidae)[J]. Arthropod Structure & Development, 2020, 55: 100926.
Tong W , Li X , Wang E , et al. Genomic insight into the origins and evolution of symbiosis genes in Phaseolus vulgaris microsymbionts[J]. BMC Genomics, 2020, 21.
LI X , WANG H , TONG W , et al. Exploring the evolutionary dynamics of Rhizobium plasmids through bipartite network analysis[J]. Environmental Microbiology, 2020, 22(3): 934–951.
Yu H , Meng W , Mao Y , et al. Deciphering the rules of mRNA structure differentiation in Saccharomyces cerevisiae in vivo and in vitro with deep neural networks[J]. RNA Biology, 2019, 16(1):1-11.
Yao M , Rahman S U , Wang A , et al. Evolutionary Analysis of the F-Box Gene Family in Saccharomycetaceae[J]. DNA and Cell Biology, 2019, 38(4).
Li F , He J , Yi Q , et al. Rapid Identification of Major QTLS Associated With Near- Freezing Temperature Tolerance in Saccharomyces cerevisiae[J]. Frontiers in Microbiology, 2018, 9:2110-.
Hua Y , Tao S H , Hua B Z . An enigmatic new species of Panorpa Linneaus from the Bashan Mountains (Mecoptera, Panorpidae)[J]. ZooKeys, 2018, 777(777):109-118.
Tong W , Li X , Huo Y , et al. Genomic insight into the taxonomy of Rhizobium genospecies that nodulate Phaseolus vulgaris.[J]. Systematic & Applied Microbiology, 2018:S0723202018300924.
Rahman, Siddiq, Ur, et al. Analysis of codon usage bias of Crimean-Congo hemorrhagic fever virus and its adaptation to hosts[J]. Infection Genetics & Evolution Journal of Molecular, 2018.
Xiangchen L , Wenjun T , Lina W , et al. A Novel Strategy for Detecting Recent Horizontal Gene Transfer and Its Application to Rhizobium Strains.[J]. Frontiers in Microbiology, 2018, 9:973-.
Ur Rahman S , Mao Y , Tao S . Codon usage bias and evolutionary analyses of Zika virus genomes[J]. Genes & Genomics, 2017.
Xianwen, Meng, Chen, et al. Genome-wide identification and evolution of HECT genes in soybean.[J]. International journal of molecular sciences, 2015.
CHANG Y , HUA Y , JIANG X , et al. Influences of Dominance and Evolution of Sex in Finite Diploid Populations[J]. GALLI A. PLOS ONE, 2015, 10(5): e0128459.
Tao S , Cheng J , Pang H , et al. Neighboring-Site Effects of Amino Acid Substitutions in the Mouse Genome[J]. Current Bioinformatics, 2015, 10(3):-.
Ailan W , Mingchuan F , Xiaoqian J , et al. Evolution of the F-Box Gene Family in Euarchontoglires: Gene Number Variation and Selection Patterns[J]. Plos One, 2014, 9(4):e94899.
Mingchuan F , Zhuoran H , Yuanhui M , et al. Neighbor Preferences of Amino Acids and Context-Dependent Effects of Amino Acid Substitutions in Human, Mouse, and Dog[J]. International Journal of Molecular ences, 2014, 15(9):15963-15980.
Yuanhui M , Huiling L , Yanlin L , et al. Deciphering the rules by which dynamics of mRNA secondary structure affect translation efficiency in Saccharomyces cerevisiae[J]. Nuclc Acids Research, 2014(8):4813-22.
Jian C , Zhao X , Wenwu W , et al. Training Set Selection for the Prediction of Essential Genes[J]. Plos One, 2014, 9(1):e86805.
Cheng J , Wu W , Zhang Y , et al. A new computational strategy for predicting essential genes[J]. BMC Genomics, 2013, 14.
Mao Y , Wang W , Cheng N , et al. Universally increased mRNA stability downstream of the translation initiation site in eukaryotes and prokaryotes[J]. Gene, 2013, 517(2):230-235.
Yuanhui M , Qian L , Wangtian W , et al. Number Variation of High Stability Regions Is Correlated with Gene Functions[J]. Genome Biology and Evolution,5,3(2013-2-13), 2013(3):484-493.
Junjie Z , Jingshi Z , Ping Z , et al. A Meta-Analysis of the Association between the hOGG1 Ser326Cys Polymorphism and the Risk of Esophageal Squamous Cell Carcinoma[J]. Plos One, 2013, 8(6):e65742.
JIANG X , HU S , XU Q , et al. Relative effects of segregation and recombination on the evolution of sex in finite diploid populations[J]. Heredity, 2013, 111(6): 505–512.
Nan C , Yuanhui M , Youyi S , et al. Coevolution in RNA Molecules Driven by Selective Constraints: Evidence from 5S rRNA[J]. Plos One, 2012, 7(9):e44376.
WU W , HUANG X , CHENG J , et al. Conservation and Evolution in and among SRF- and MEF2-Type MADS Domains and Their Binding Sites[J]. Molecular Biology and Evolution, 2011, 28(1): 501–511.
Jiang X , Xu Z , Li J , et al. The Influence of Deleterious Mutations on Adaptation in Asexual Populations[J]. PLoS ONE, 2011, 6(11):e27757-.
Wenwu W , De F S , Xia S , et al. Vertebrate Paralogous MEF2 Genes: Origin, Conservation, and Evolution[J]. Plos One, 2011, 6(3):e17334.
Pang H , Tang J , Chen S S , et al. Statistical distributions of optimal global alignment scores of random protein sequences[J]. BMC Bioinformatics, 2005, 6(1):1-9.
实验室网页
实验室风采





